Looking back | How we used supercomputers to fight COVID-19
Meet Ya-Wen Hsiao, a Computational Scientist at the Hartree Centre, part of a team who worked with IBM Research and used molecular dynamics simulations to map mutations in COVID-19 as part of the pandemic response.

Your group has recently published a paper about your research into SARS-CoV-2/hACE2 (COVID-19) and how its structure adapts. For people who are unfamiliar with COVID-19 can you please explain how the virus works?
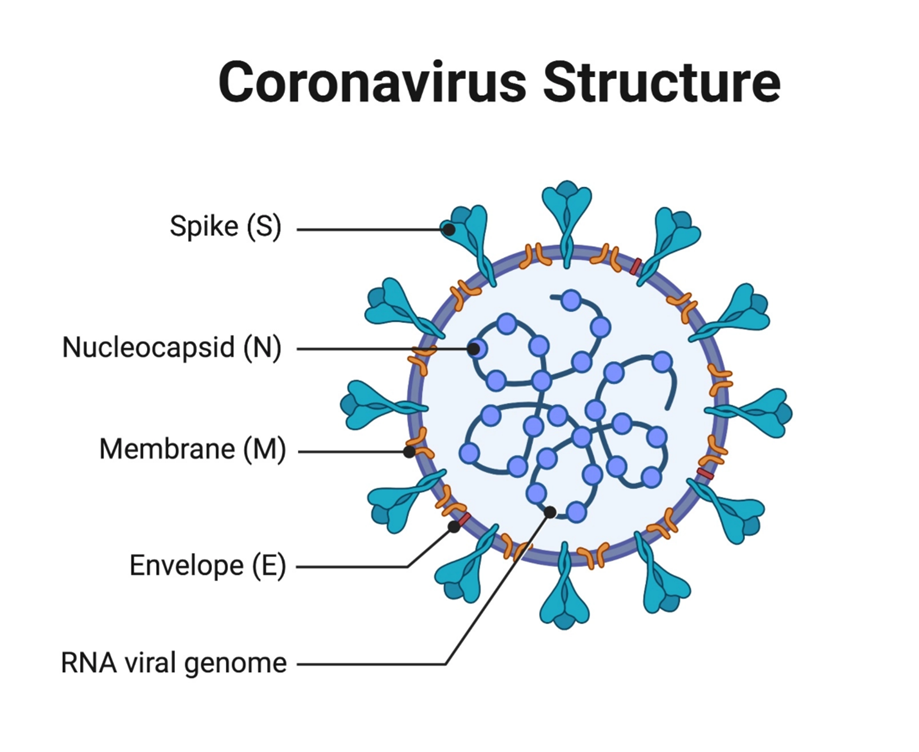
COVID-19 is a member of the coronavirus family, which causes a respiratory illness. There have been over 500 million cases worldwide since the start of the global pandemic in 2020. COVID-19 works by using the proteins in the virus to attach to receptors on healthy cells, especially in lungs. Part of the reason why it has spread so rapidly and has impacted so many people is the way the virus has mutated to adapt the structure of its spike protein.
Several new variants of COVID-19 developed since the start of the pandemic through several mutations. The World Health Organization has designated Alpha, Delta and Omicron as variants of concern because of their increasing transmission rate and how viral they are.
In our research, we focused on the structural adaptation in the Omicron variant, as the mutation in this strain made it the dominant form of the virus. When tackling viruses, it is important to appreciate that they interact differently so that we can use that understanding to create targeted treatments. Going forwards, we have applied what we have learned in this research to help develop other ways to map molecules, which can be used to create better medical treatments.
You have mentioned the mutations in the Omicron variant of the COVID-19 virus, why was this important to research and what were you looking for?
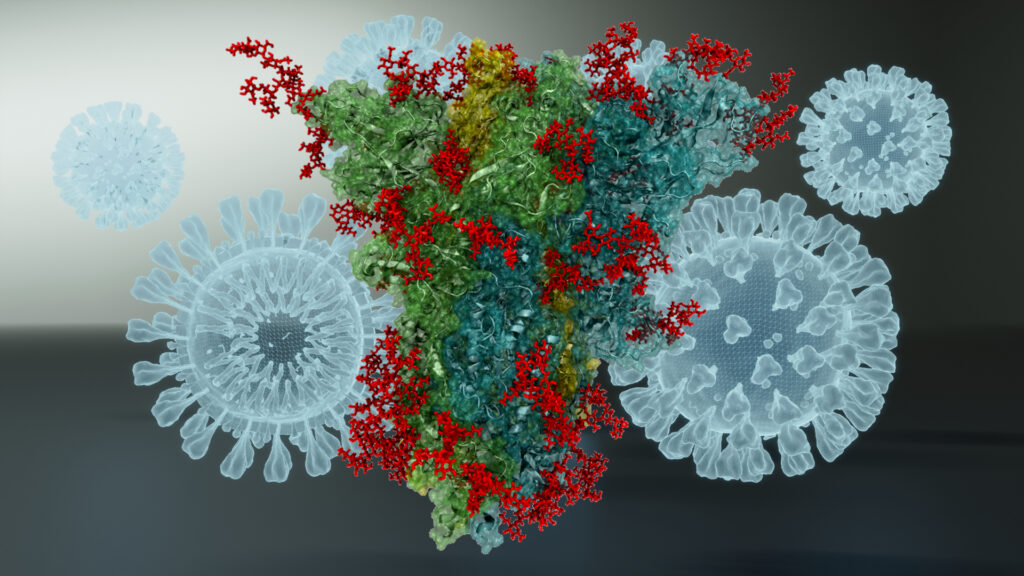
In our research we studied the spike protein, the part of the virus that attacks the body by latching onto the healthy cell receptors in the lungs. As the dominant strain of the virus, Omicron is unique among known variants as the spike protein has mutated over thirty times. It is important to learn how the spike protein functions to understand how the virus can attack the body. Using supercomputers, we studied the structure of the Omicron variant to understand how it mutated and adapted to increase its transmission.
So these mutations to the spike protein structure are what made the Omicron variant spread so rapidly, how were you able to analyse these adaptions?
We used molecular dynamic (MD) simulations to visualise the structure of the Omicron variant to analyse how it muted over thirty times. This helps pick apart the infection process and the molecular mechanisms of the COVID-19 receptor-binding domain (RBD) located on the spike protein which is what recognises and latches onto the healthy cell receptors in the lung. These simulations were then run on the Hartree Centre’s supercomputer, Scafell Pike, enabling fast solve times and detailed visual models of the protein. My group were able to model several subvariants of Omicron from our simulations, finding a part of what made this variant of the virus become so transmissible and the dominant strain of COVID-19 during the pandemic.
From what you are describing, visualising COVID-19 helped change your understanding of the mutations. What does this mean for analysing variants of the virus and how does this impact the way we view it moving forward?
Well one of the things we are looking at is further mutation of the virus, as the vaccine strategy relies heavily on the spike sequence and the neutralising ability of vaccine-induced antibodies. There are concerns that a hypermutated strain could emerge and bypass these antibodies. There has been some evidence of this in the subvariants of Omicron in those who are only double-vaccinated, but this is overcome when a booster dose is administered. We traced this change to a protein mutation, which is why this sort of research is so important as you can simulate and visualise the molecules. Giving scientists the ability to map structural differences that could explain why this variant of the virus became more transmissible than earlier forms. We are now using what we have learned through this research in current projects to explore different ways we can map molecules, helping provide a new perspective.
Join Newsletter
Provide your details to receive regular updates from the STFC Hartree Centre.